Metabolic Perceptrons for Neural Computing in Biological Systems, Nature Communications
While machine learning methods are used in many areas, including human health, interfacing these methods with the living world has been little explored at the molecular level. We have just taken a first step in this direction by engineering a simple neural network in an E. coli cell-free extract. The method enables one to classify samples with different metabolites concentrations. The results are published in Nature Communications1.
We have witnessed the past decades tremendous progress made in machine learning including deep learning recently. Machine learning is now used in many tasks that affect our daily life and is currently making strives regarding our health. Yet, little has been done to interface machine learning with biological systems at the molecular level, this is noteworthy considering that deep learning methods are inspired by biological systems, i.e., the neural network of our brain.
Even though chemical and biological systems can perform computations, their performances lack far behindin silicocomputing power. When coupling machine learning with biological systems, it is therefore more efficient to still carry out computation in silico, while acquiring computational input and actuating output in vitroor in vivo.
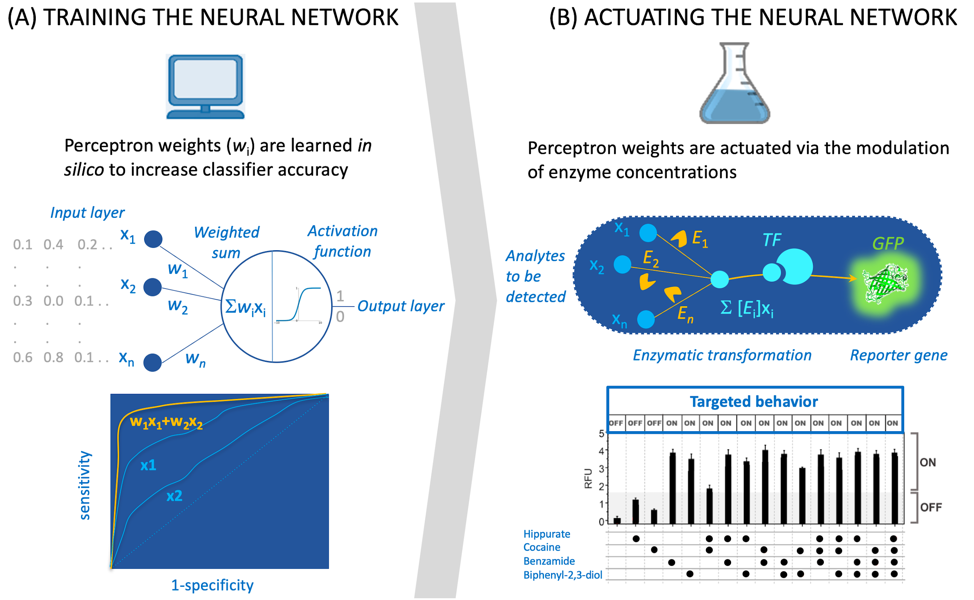
Taking medical diagnostic as an application example and focusing on neural networks (Figure 1), one could conceive a system where the network weights are learned in silicoduring a training phase, and the network output (yes or no for a diagnostic) is actuated by an engineered biological system. For diagnostic purposes, input training data could be composed of experimental biomarker concentrations measured on many samples for specific diseases. As any machine learning tool, such a system could be used to predict the outcome for new samples that have not been seems during the training phase, and ultimately be used as a point of care testing (POCT) device for diagnosis.
Learning neural network weights for diagnosis using biomarker concentrations has already been undertaken for several diseases2. Previous studies have been carried out in silicoand require omics data to both train the network and used it to diagnose. Taking actual clinical samples as network input and producing an output that can be directly and easily measured requires to actuate the network into an engineered biological system. As a first step toward this endeavor, we engineered a simple neural network that can perform classification based on the concentrations of input metabolites1. In our work, the network weights are learned in silicoand actuated in an E. colicell-free system thought the modulation of enzyme concentrations (Figure 1).
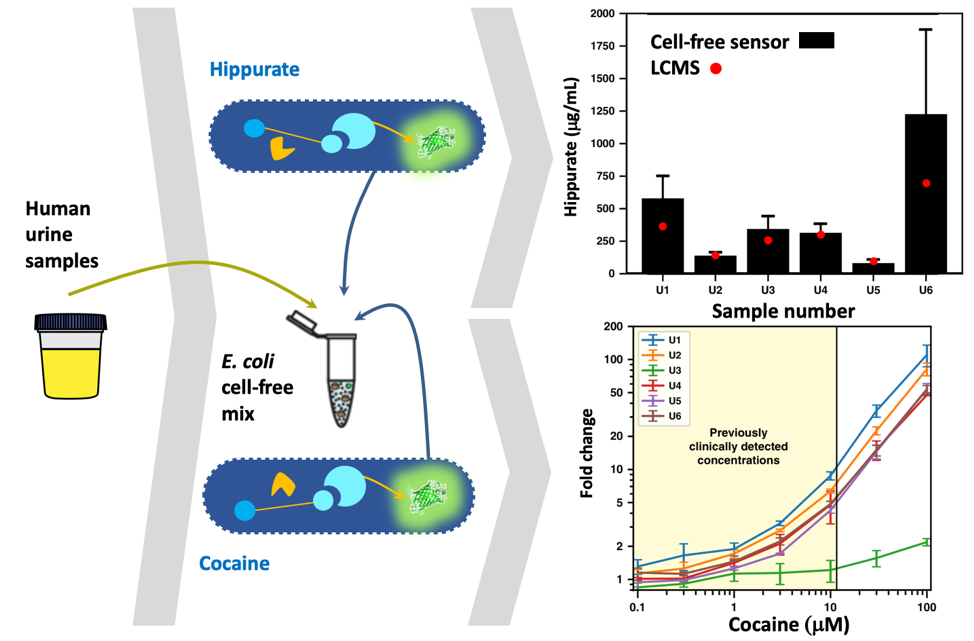
To probe the ability of our engineered systems to work on real world samples we partnered with J. Bonnet’s group at CBS (INSERM, Montpellier) and performed validation tests on human urine for metabolites detection (Figure 2) in a second study also published in Nature Communications3. Among the selected metabolites, cocaine was picked for obvious applications in drug law enforcement, and hippurate was chosen because it can be used to monitor a recent probiotic treatment of phenylketonuria. Phenylketonuria is a neurotoxic disease characterized by the inability to process phenylalanine. Isabella et al.4 have recently shown that adding a lyase to E. coli Nissle transforms phenylalanine into trans-cinnamate, which in turn is metabolized into hippurate by the patient’s liver. Thus, engineering a low-cost device based on E. coli cell extract enables one to monitor treatment efficacy avoiding more expensive and time-consuming mass spectrometry measurements.
Figure 1. Interfacing in siliconeural computations with biological systems. (A) The simple network is a one-layer perceptron. It can be used for classification (output is 0 or 1) provided data is fed to the system (in gray). During training, weights (wi) are calculated in order to maximize the area under the ROC curve (bottom). (B) The network represented in (A) is implemented via enzymatic transformations that metabolize analytes (the system input) into a transcription factor (TF) activator, the activated TF triggers the expression of a reporter gene (green fluorescent protein GFP). The graph at the bottom plots the fluorescence measurements (RFU: Relative Fluorescence Unit) of a 4-input perceptron (analytes/inputs are hippurate, cocaine, benzamide and biphenyl-2,3-diol). Results matches well the targeted behavior.
Figure 2. Benchmarking with real world samples. Both hippurate and cocaine are enzymatically transformed (yellow) into benzoate, which activates the transcription factor BenR (light blue), triggering the expression of a reporter gene (green). The two systems engineered in an E. colicell-free mix are used to quantify metabolites in human urine sample. For hippurate, the cell-free sensor reports concentration similar to those obtained with a more traditional Liquid-Chromatography Mass Spectrometry (LCMS) techniques. For cocaine the dynamic range of the cell-free sensor is in the range of clinically detected concentrations. Fold change stands for fluorescence fold change.
References
- Pandi A, Koch M, Voyvodic PL, Soudier P, Bonnet J, Kushwaha M, Faulon JL. Metabolic Perceptrons for Neural Computing in Biological Systems. Nature Communications, 10: 3880, 2019. | doi: 10.1038/s41467-019-11889-0
- Zafeiris D, Rutella S, Ball GR. An Artificial Neural Network Integrated Pipeline for Biomarker Discovery Using Alzheimer’s Disease as a Case Study. Computational and Structural Biotechnology Journal, 16: 77–87, 2018. | doi: 10.1016/j.csbj.2018.02.001
- Voyvodic PL, Pandi A, Koch M, Conejero I, Valjent E, Courtet P, Renard E, Faulon JL, Bonnet J. Plug-and-play metabolic transducers expand the chemical detection space of cell-free biosensors. Nature Communications, 10: 1697, 2019. | doi: 10.1038/s41467-019-09722-9
- Isabella VM, Ha BH, Castillo MJ, Lubkowicz DJ, Rowe SE, Millet YA, Anderson CL, Li N, Fisher AB, West KA, Reeder PJ, Momin MM, Bergeron CG, Guilmain SE, Miller PF, Kurtz CB, Falb D. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nature Biotechnology, 36: 857–864, 2018.