Can we compute with metabolism?
Keywords: Systems Biology, Deep Learning, Synthetic Biology
Project Summary:
Can we engineer a perceptron (basic block of neural network architecture) in a microorganism? Conversely, can we use cellular network motifs as architectures for deep neural network?
Prior answering these questions, we first need to probe if and which information processing modules exist in the natural world and in particular in microorganisms we can engineer. This is the main goal on the internship. To search for natural motifs and motifs that can be engineered, the student will benefit from the team expertise in metabolism and information processing we have developed the past few years [1,3,5].
Description
Quorum molecules, hormones, neurotransmitters and extracellular chemicals produce signals that are transduced down to the genetic layer in living organisms. Information is generally passed on through cellular receptors then signaling pathways to ultimately produce a transcriptional response.
In most instances, transcriptional activation requires the multiplexed detection and integration of different signals. For instance, bacteria like Pseudomonas aeruginosa are capable of detecting multiple quorum molecules to regulate virulence and biofilm formation through multi-layered signaling networks [2a]. More sophisticated are the complex decision-making molecular circuits (similar to perceptron) found in plants to control responses to a large range of environmental stimuli, interconnected together to what has been named the wood wide web [2b].
Enzymes are key players in signal transduction as they are the catalyst of the pathways. Since enzyme produce metabolites, there are also instances in which signals are propagated through metabolism [3]. The best-known example is within the lac operon, where the transcription factor LacI is activated by allolactose, a metabolite produced from lactose by a secondary activity of beta-galactosidase. The underground activity of 𝛽-Galactosidase is present only when LacI is present and this network motif is conserved by evolution [3a].
The goal of the internship will be to probe to which extent natural metabolism plays a role in transducing, integrating and processing signals in particular with bacteria where signaling pathways are not as developed as in higher organisms.
The work will consist of searching motifs in metabolic databases corresponding to signal transduction, integration and processing. For signal transduction one will have to search for pathways linking metabolites to transcription factors or riboswitches, for signal integration and processing one might consider logic gates as it has been suggested in the past [4a] or analog electronic motifs such as (weighted) adder, subtractor, attenuator, perceptron, and recurrent loop, some which already found in signaling pathways [4b]. To validate the motifs found in metabolic maps of specific organisms, the work should yield experimental designs to be carried by the team members of the recruiting group and/or during a follow-on PhD, who have experience in engineering metabolic circuits [5a-d].
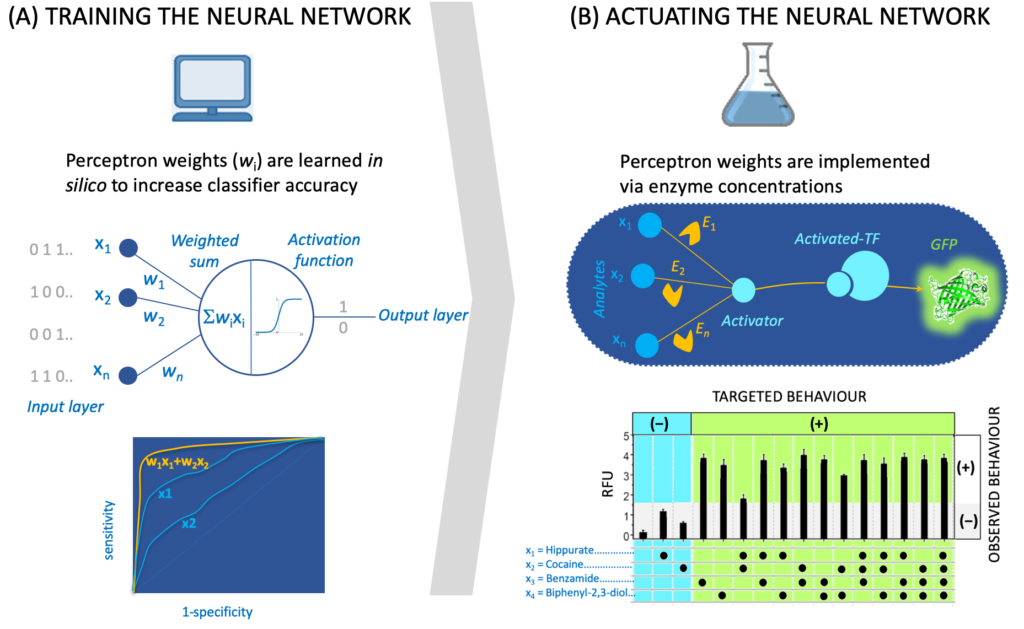
The recruited student will benefit from preliminary work we have carried out exhibiting examples beyond allolactase and LacI where metabolic reactions are transducing signals [3a-b, 5a-d] along with a database [1c] and a software tool (SensiPath [1b]) we have compiled for sensing enabling metabolic pathways. The recruited student will work closely with an IT engineer (Thomas Duigou), a research associate (M. du Lac) and a PhD student (Paul Soudier) who are currently designing and engineering signal transduction, integration and processing using metabolic pathways in the context of whole cell and cell-free biosensors.
Possible Work plan outline for a 6-month internship
- Task 1. Literature survey and motif search code benchmarking (0.5 month). Review literature on motif detection (applicable to metabolic networks). Benchmark a code written by a former master student (Ivan Valiev, Skoltech Institute, Moscov) to search for motifs in metabolic networks (using Kegg maps for instance).
- Task 2. SensiPath Database update (0.5 months). You will learn the tools we have developed to build the database (mostly the RetroPath2.0 [1a] workflows available at MyExperiment.org), you’ll then generate an updated database with these tools.
- Task 3. Motif search in updated SensiPath database (4 months). Here you will have first to work out the topology of the motifs to be searched. Potential motifs are transducer, actuator, adders, 1 layer or multilayer perceptrons, recurrent loops, bi-fan, and logic gates. Once the motifs will have been designed you will search them on the database build in Task 2 and with the tools developed in Task 1. Most of the work here will be to determine the topology of the motifs, as the search itself should be straightforward. You will also propose plan of experiments for experimental validation of the motifs found on SensiPath Database. There will be a possibility to carry out experimental work during a follow-on PhD.
- Task 4. Complete the work started in Task 3 and write report suitable for publication (1 month).
Références (de l’équipe *) sur le sujet
1* (a) Delepine, B., et al., RetroPath2.0: A retrosynthesis workflow for metabolic engineers. Metab Eng, 2018. 45: 158-170. (b) Delepine, B., et al., SensiPath: computer-aided design of sensing-enabling metabolic pathways. Nucleic Acids Res, 2016. 44(W1): W226-31. (c) Koch, M., et al., A dataset of small molecules triggering transcriptional and translational cellular responses. Data Brief, 2018. 17: 1374-1378. (d) Duigou T. et al. RetroRules: a database of reaction rules for engineering biology. Nucleic Acids Research, 47(D1): D1229-1235, 2019
2 (a) Lee, J., et al., The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell, 2015. 6(1): 26-41. (b) Scheres, B., et al., The plant perceptron connects environment to development. Nature, 2017. 543(7645): 337-345.
3* (a) Libis, V., et al., Sensing new chemicals with bacterial transcription factors. Current Opinion in Microbiol, 2016. 33: 105-112. (b) Koch M., et al. Custom-made transcriptional biosensors for metabolic engineering. Current Opinion in Biotechnology, 2019, 59:78-84.
4 (a) Arkin, A., et al., Computational functions in biochemical reaction networks. Biophys J, 1994. 67(2): 560-78. (b) Alon, U., An Introduction to Systems Biology: Design Principles of Biological Circuits. 2006: Chapman and Hall/CRC.
5* (a) Libis, V., et al., Expanding Biosensing Abilities through Computer-Aided Design of Metabolic Pathways. ACS Synth Biol, 2016. 5(10): 1076–1085. (b) Pandi A, et al. Optimizing Cell-Free Biosensors to Monitor Enzymatic Production. ACS Synth Biol. 2019, 16;8(8):1952-1957. (c) Voyvodic PL, Pandi A, et al., Plug-and-play metabolic transducers expand the chemical detection space of cell-free biosensors. Nature Communications, 10(1):1697, 2019. (d) Pandit A. et al. Metabolic Perceptrons for Neural Computing in Biological Systems. Nature Communications 2019, Nature Communications 10, Article number: 3880 (2019)



