BioRetroSynth: background and previous developments
Our aim is to demonstrate that retrosynthesis, a widely used technique in chemistry, can be utilized to engineer chassis organisms such as E. coli to biosynthesize specific target compounds. Retrosynthesis analysis transforms a synthetic target product into precursors, following pathways to commercially available starting materials. The transformations are applied in the reverse direction from the actual synthesis. In the case of metabolic engineering, retrosynthesis consists of applying reversed biotransformations (i.e., reversed enzymes catalyzed reactions) to a target product, following pathways to substrates that are endogenous to a chassis organism.
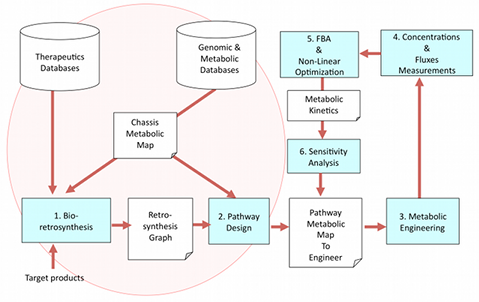
Bioretrosynthesis. The algorithm of retrosynthesis that we have developed searches for heterologous genes and pathways, which once introduced into a given chassis organism can produce a target compound. These heterologous genes and their associated metabolites are organized into an annotated retrosynthesis graph where substrates, products and reactions are coded into molecular signatures. We use machine learning to mine genomic databases for predicting protein function and enzymes catalyzing specific substrates. We are currently using the algorithm to build retrosynthesis graphs for heterologous antibiotics production in E. coli.
Pathway design. Enumerate and rank all possible pathways in the retrosynthesis graphs to help decide which pathways are best to engineer to produce a given target product. Enzymes can potentially process multiple substrates or reactions, we study enzyme promiscuity to enhance enzyme efficiency by protein engineering techniques. In addition, we have developed a Quantitative Structure-Activity Relationship (QSAR) for enzyme activity and inhibition based on experimental databases and toxicity assays. This QSAR is used to reverse engineer enzymes having specific activity through a process named inverse QSAR.
Metabolic engineering. We engineer E. coli plasmids in order to construct combinatorial libraries of highest rank heterologous pathways found to produce a target product. Combinatorial libraries are generated through the use of assembly PCR tested first in vitro, then in vivo. Production of the target metabolite is verified by measuring metabolites concentrations and fluxes.
Engineering optimization. We use Flux Balance Analysis (FBA) and non-linear optimization methods to populate the metabolic map of the engineered chassis with kinetics parameters and to determine positive and negative enzyme regulations to maximize target yield. The robustness of the optimal fluxes are sampled in order to perform a sensitivity analysis.
Please visit also our web server for retrosynthetic metabolic pathway design.



